Glowing Recommendations
Fluorescent proteins are the tiny, glowing workhorses of biomedical imaging.
When researchers want to observe the activity of a gene in a cell, an organ or a whole organism, they can insert a short DNA sequence that codes for fluorescence into the gene. The protein made by that gene then glows under fluorescent light.
“When we want to study a protein and see where it goes over time and space, we can’t usually see it directly,” explained Talley Lambert, manager of the Cell Biology Microscopy Facility at HMS. “One of the main tools we use are these fluorescent proteins that absorb and emit light.”
Since green fluorescent protein (GFP), originally discovered in jellyfish, became the first of these tools adapted for scientific research in the 1990s, earning a Nobel Prize in 2008, dozens upon dozens of fluorescent proteins have flooded the market in a rainbow of colors and a range of other essential properties.
Now researchers designing fluorescence microscopy experiments are spoiled for choice—and the breadth of options creates a conundrum: Which proteins are best suited to their projects?
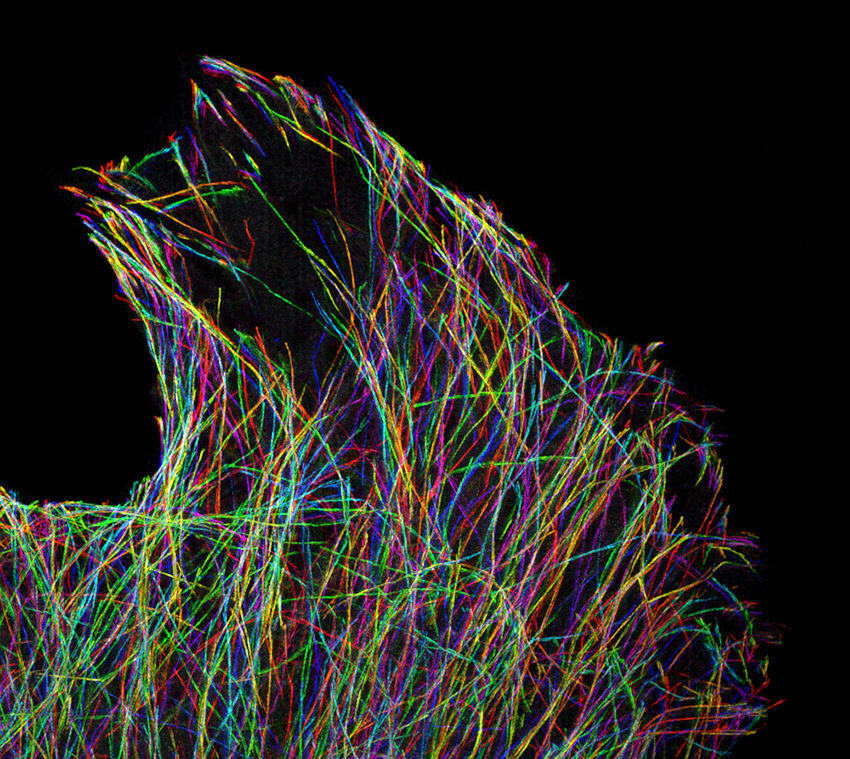
Fluorescent proteins come in hues with names such as dTomato, Kusabira Orange, mBanana, Emerald, and Azurite. Image: Cell Biology Microscopy Facility
“It’s an extremely common question,” said Lambert, who is also a research associate in cell biology at HMS. “You want to know which is right before you spend three weeks fusing the protein to your gene of interest.”
The wrong choice can hamper experiments or confound results. However, researchers don’t necessarily have the training to understand all the factors that distinguish one fluorescent protein from another. Compounding the problem, there has been no central repository for those who wanted to look up such details.
That has now changed.
One-stop shopping
To help researchers at HMS and beyond identify the proteins that will best achieve their goals, Lambert has built the Fluorescent Protein Database, or FPbase.
The searchable, web-based tool aggregates information about fluorescent proteins from multiple sources and delivers them in one place in user-friendly formats.
Using FPbase, researchers can explore—and set constraints for—the spectra that fluorescent proteins absorb and emit as well as their brightness, acid sensitivity, photostability (how long a protein can withstand light exposure), quantum yield (how efficiently it converts absorbed light into emitted light), lifetime in nanoseconds, and more. Each data set provides references to original publications about the proteins.
FPbase also includes a feature that allows researchers to enter key attributes of the microscopes in their labs, such as lasers, filter sets, and cameras, to help them determine how well different fluorescent proteins will work with their particular instruments, says Jennifer Waters, lecturer on cell biology and director of microscopy in the Department of Cell Biology at HMS.
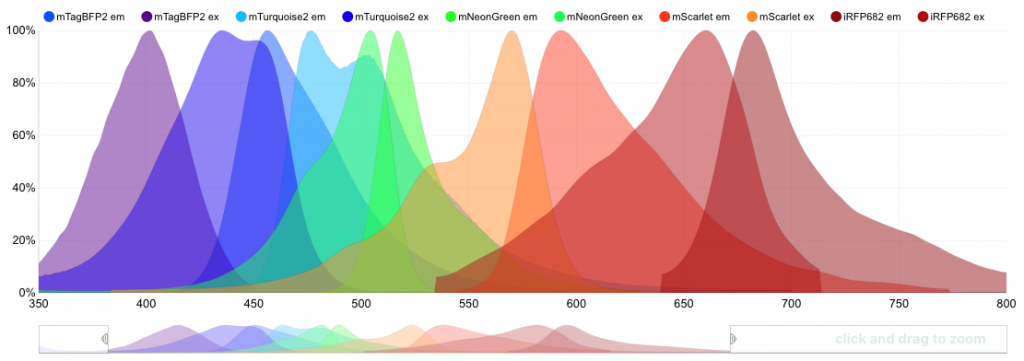
FPbase allows researchers to easily compare the absorption and emission spectra of fluorescent proteins. Image: Talley Lambert
In addition to its comprehensiveness and usability, FPbase is the only fluorescent protein database designed to be editable by community members, Wikipedia-style.
“Users can upload information into the database, allowing it to grow and stay current without any one person or group needing to maintain it,” Waters explained.
“If something isn’t satisfying, you can make that part of the website better for you and others,” added Lambert. “You can update and edit if you find mistakes.”
Lambert moderates user-contributed content to ensure that people can add to FPbase without “messing it up,” he says. Users must provide citations, and he can make corrections or fill in missing information.
“I hope people use FPbase not just to consume information but to contribute,” he said. “I want it to take on a life of its own.”
Related story: Dream Machines
Previous
Image: Cell Biology Microscopy Facility
Image: Cell Biology Microscopy Facility
1/4
Image: Cell Biology Microscopy Facility
1/4
Image: Cell Biology Microscopy Facility
1/4
Image: Cell Biology Microscopy Facility
1/4
Image: Cell Biology Microscopy Facility
Image: Cell Biology Microscopy Facility
Image: Cell Biology Microscopy Facility
Image: Cell Biology Microscopy Facility
Next
Hiding in plain sight
Beyond its main purpose, FPbase has begun providing a foundation for new knowledge.
A group that makes fluorescent proteins cited FPbase in a recent study. Building on a graph generated by the database, the researchers discovered that two properties of fluorescent proteins—quantum yield and lifetime—are correlated.
“The more data you get, the more patterns you see,” said Lambert. “As FPbase grows, people can potentially generate new observations and intuitions.”
In particular, Lambert hopes FPbase will inspire hypotheses that can guide the design of new fluorescent proteins.