Small Babies, Big Data
Technique reveals detailed developmental trajectory during first week of life
The first week of a newborn’s life is a time of rapid biological change as the baby adapts to living outside the womb, suddenly exposed to new bacteria and viruses. Yet surprisingly little is known about these early changes.
An international research team co-led by Harvard Medical School researchers at Boston Children’s Hospital has pioneered a new technique to get vast quantities of data from a tiny amount of newborn blood, creating the most detailed account to date of molecular changes in the first week of life.
As reported March 12 in Nature Communications , the data includes what genes are turned on, what proteins are being made and what metabolites are changing.
By establishing a common developmental pathway for the first week of life, the work provides a baseline to further our understanding of newborn health and, in particular, the impacts of vaccines on newborns.
“Most infections in the world occur early in life, and newborns have the greatest susceptibility and the worst outcomes,” said Ofer Levy, HMS professor of pediatrics, director of the Precision Vaccines Program at Boston Children’s and a senior author on the paper. “This work provides a valuable window into health and disease in the first week of life.”
The team included members of the Expanded Program on Immunization Consortium (EPIC), an international collaboration that employs a systems biology approach to understand vaccine action in early life.
Levy partnered with Beate Kampmann of the Medical Research Council Unit The Gambia at the London School of Hygiene & Tropical Medicine and Tobias Kollmann of the University of British Columbia (UBC) to establish the consortium, which has its administrative hub at Boston Children’s.
Tiny sample, big knowledge
Previous efforts to gather data on newborn development were limited by the challenge of obtaining a large enough blood sample from a tiny newborn.
The team overcame this challenge with new techniques, partnering with Robert Hancock of UBC, who led an RNA analysis, and Hanno Steen, HMS associate professor of pathology at Boston Children’s, who led a proteomics analysis.
Using sophisticated software to analyze complex data derived from less than half a teaspoon of blood, the UBC labs of Hancock and Scott Tebbutt found thousands of changes over the first week, including changes in gene expression and components of immune defense such as interferons, neutrophil function and complement pathways.
“We demonstrated that it’s possible to recruit newborns in a resource-poor setting, obtain small amounts of their blood, process it, ship it, conduct systems biology assays and integrate the results—turning big data into knowledge,” said Levy.
Key to the analysis, Levy said, was comparing each newborn at day one, day three and day seven to their own baseline condition on the day of birth.
“That’s when we discovered dramatic molecular changes driven by development,” he said.
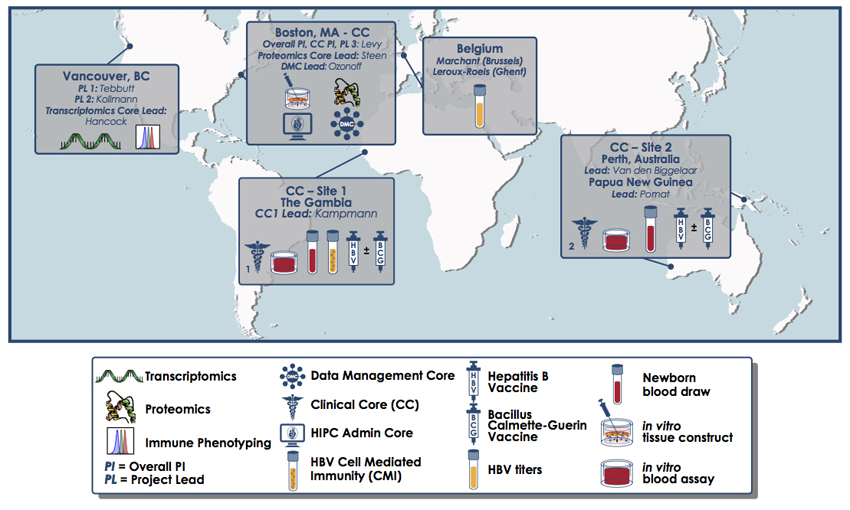
Study collaborators were located in the United States, Canada, Belgium, Gambia, Australia and Papua New Guinea. Image: Kristin Johnson/Boston Children’s
Intercontinental effort
The researchers piloted their systems-biology approach with a group of infants from Gambia in West Africa after first obtaining permission from village elders and informed consent from mothers in local languages such as Mandinka, Fula and Wolof.
They then validated their approach with a second group of Australasian newborns recruited by William Pomat of the Papua New Guinea Institute of Medical Research and Anita van den Biggelaar of the University of Western Australia.
The two independent cohorts showed a common, highly dynamic developmental trajectory. The findings suggest that the changes do not occur at random but instead follow an age-specific pathway.
“This common trajectory is exciting as it allows us to ask bigger questions about the differences between different populations and the impact of biomedical interventions such as vaccines on development,” said Levy.
Optimizing vaccines in early life
The study establishes a baseline for health and disease in early life that can help measure responses to key medical interventions and the impact of factors such as diet, disease and maternal health.
Levy is particularly interested in studying the impact of immunization. Newborns’ responses to immunization are distinct from those of older individuals, and much needs to be learned to optimize the use and benefit of vaccines in early life.
Currently, most vaccines are developed by trial and error, said Levy. “We seek deep molecular insight into vaccine function in early life so we can better develop infant vaccines for the future,” he said.
Funding and authorship
The study was supported in part by the National Institutes of Health’s National Institute of Allergy and Infectious Diseases as part of the Human Immunology Project Consortium (5U19AI118608-02) and by the Precision Vaccines Program.
Co-first authors were Amy Lee, Casey Shannon and Nelly Amenyogbe of UBC; Tue Bennike of Aalborg University, Denmark; Joann Diray-Arce, HMS research fellow in pediatrics at Boston Children’s; and Olubukola “Bukky” Idoko of MRC-Gambia/LSHTM. Co-senior authors were van den Biggelaar, Steen, Tebbutt, Kampmann, Levy and Kollmann